There is a lot of code that needs to make a selection based on a maximum value. One example is a Kafka read: we only want the latest offset for each key, because that's the latest record. What is the fastest way of doing this?
Selecting the max value
I've seen two ways of doing this. The first way creates a new dataframe with the maximum value and the key and joins it back on the original dataframe, so other values are filtered out. The second way uses an aggregation and a struct-column that has the max value as the first column of that struct.
Both wave give the same result, but which one is faster? To make a comparison, I've created a dataframe one of our product delta tables with 943.546 records to see what happens in terms of performance when we select the maximum product number per release date. The number of products number it should find is 7.817.
Max by join
Our first method uses a "join dataframe". In this dataframe we'll group by the release date and determine the max product number. We'll join it back on the original dataframe and count the number of records (so an action is triggered). We'll return the time it took.
def max_by_join(df_source):
start = time.time()
df_top = (df_source
.select("releaseDate", "productNumber")
.groupBy("releaseDate")
.agg(max("productNumber").alias("productNumber"))
.select("productNumber"))
df_final = df_source.join(df_top, ["productNumber"])
print(df_final.count())
diff = time.time() - start
return diffMax by agg / struct
Our second method does almost the same, but before aggregating on the release date, it will make a selection of the release date and a special struct. The first field of this struct is the product number as mx and the rest of the struct is the rest of the record. The first product number is renamed, because otherwise the column name would be a duplicate.
The dataframe is then grouped by release date and a max is done over the struct. Because the first field of the struct is the product number, only the max records are kept. Now all the fields of the struct are selected and the mx field is dropped. This function also returns the time it took.
def max_by_agg(df_source):
start = time.time()
df1 = (df_source
.select("releaseDate", expr("struct(productNumber as mx, *) as r"))
.groupBy("releaseDate")
.agg(max("r").alias("r"))
.select("r.*")
.drop("mx"))
print(df1.count())
diff = time.time() - start
return diffMinimizing effects
To minimize the effect of the first read of the dataframe, I did a df_source.count() before starting the loop.
The order of execution might also influence the result. So we're using a int(bool(random.getrandbits(1))) to determine which function should go first. 0 is the max_by_join and 1 the max_by_agg. The order is determined for every run.
We're also using a Delta table, so it might choose to do some updates while we're doing our simulation.
We'll run 30 times.
Results
I've plotted the results in this table:
+-----+--------------+--------------+--------+--------+
| Run | Join, rsi* | Agg, rsi* | First | Winner |
+-----+--------------+--------------+--------+--------+
| 1 | 2.8323, 1.43 | 1.9839, 0.70 | Agg | Agg |
| 2 | 3.2432, 1.72 | 1.8854, 0.58 | Agg | Agg |
| 3 | 3.2874, 1.57 | 2.0882, 0.64 | Agg | Agg |
| 4 | 3.5743, 1.94 | 1.8433, 0.52 | Join | Agg |
| 5 | 2.9656, 1.28 | 2.3253, 0.78 | Agg | Agg |
| 6 | 3.2783, 1.84 | 1.7790, 0.54 | Agg | Agg |
| 7 | 2.9010, 0.85 | 3.4085, 1.17 | Agg | Join |
| 8 | 3.3526, 1.76 | 1.9068, 0.57 | Join | Agg |
| 9 | 3.8894, 1.90 | 2.0501, 0.53 | Join | Agg |
| 10 | 3.1941, 1.37 | 2.3261, 0.73 | Agg | Agg |
| 11 | 3.4618, 1.80 | 1.9213, 0.55 | Join | Agg |
| 12 | 3.1267, 1.55 | 2.0230, 0.65 | Agg | Agg |
| 13 | 2.9610, 0.85 | 3.4850, 1.18 | Agg | Join |
| 14 | 3.1271, 1.57 | 1.9955, 0.64 | Join | Agg |
| 15 | 3.3198, 1.53 | 2.1733, 0.65 | Agg | Agg |
| 16 | 3.1191, 1.58 | 1.9773, 0.63 | Join | Agg |
| 17 | 3.0698, 1.68 | 1.8280, 0.60 | Join | Agg |
| 18 | 3.2186, 1.73 | 1.8611, 0.58 | Join | Agg |
| 19 | 3.0382, 1.61 | 1.8837, 0.62 | Agg | Agg |
| 20 | 2.9669, 1.51 | 1.9661, 0.66 | Join | Agg |
| 21 | 2.6517, 0.87 | 3.0421, 1.15 | Agg | Join |
| 22 | 3.2077, 1.69 | 1.8937, 0.59 | Join | Agg |
| 23 | 3.1774, 1.83 | 1.7339, 0.55 | Agg | Agg |
| 24 | 3.1343, 1.64 | 1.9159, 0.61 | Join | Agg |
| 25 | 2.6993, 1.35 | 2.0033, 0.74 | Agg | Agg |
| 26 | 2.7731, 1.35 | 2.0506, 0.74 | Join | Agg |
| 27 | 2.9683, 1.49 | 1.9913, 0.67 | Join | Agg |
| 28 | 2.8118, 1.58 | 1.7795, 0.63 | Agg | Agg |
| 29 | 2.6850, 1.39 | 1.9306, 0.72 | Agg | Agg |
| 30 | 2.7917, 1.42 | 1.9706, 0.71 | Join | Agg |
+-----+--------------+--------------+--------+--------+
rs* = relative speed index
Avg. Join: 3.0942, 1.45
Avg. Agg: 2.1230, 0.68Overall, the aggregation is faster - but not always. In cases where the join was faster, it was never executed first in the loop. It looks like the aggregation function is >30% faster.
What about 1000 runs?
I did another simulation of a 1000 runs and plotted the result in this graph:
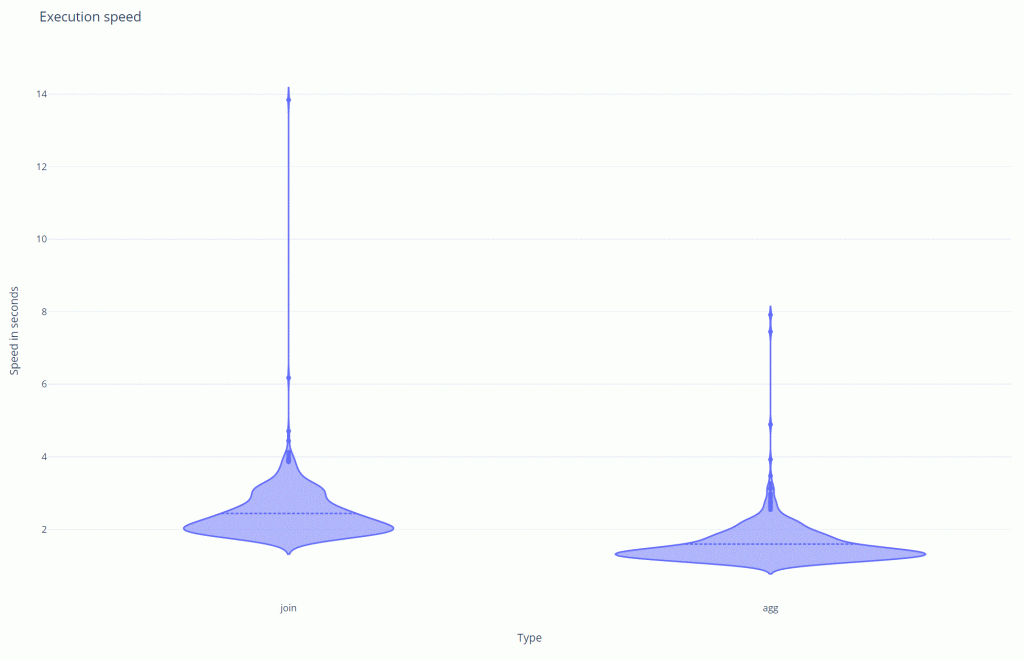
agg is.The average of the join is 2.43s vs the average of the aggregation of 1.59s. This also shows that the aggregation / struct is >30% faster.
Conclusion
The speed of queries finding maximum values can be improved by >30% when you swap out a join for an aggregation with a struct.

